Abstract
Background. Periodontitis is an inflammatory disease of the oral cavity that affects the soft and hard tissues of the periodontium due to dysbiosis by Porphyromonas gingivalis. The bacterium establishes its pathogenicity through its virulence factors, such as fimbriae, and by the releasing proteases like gingipains. The lysine-specific gingipain K (Kgp) is characterized by the presence of a hemagglutinin (HA)–adhesin domain, which provides the micronutrients for the survival of the microbe. 4-Caffeoylquinic acid (4-CQA) is classified as a phenylpropanoid. It exhibits a variety of bioactivities, including anti-inflammatory, antimicrobial, antihistaminic, and antioxidant properties. Moringa oleifera has multiple therapeutic benefits and is used in the treatment of cancer, infections, diabetes, and arthritis. 4-Caffeoylquinic acid was identified among the phenolic phytocomponents present in M. oleifera.
Objectives. The aim of the present study was to use in silico docking and a dynamic model to evaluate the potential inhibition of Lys-gingipain of P. gingivalis by 4-CQA of M. oleifera.
Material and methods. Molecular docking and dynamic simulations of the Lys-gingipain protein and 4-CQA ligands were performed using the Desmond software. The protein structure of Lys-gingipain was downloaded from the Protein Data Bank (PDB) and preprocessed using the optimized potentials for liquid simulations (OPLS 2005) force field.
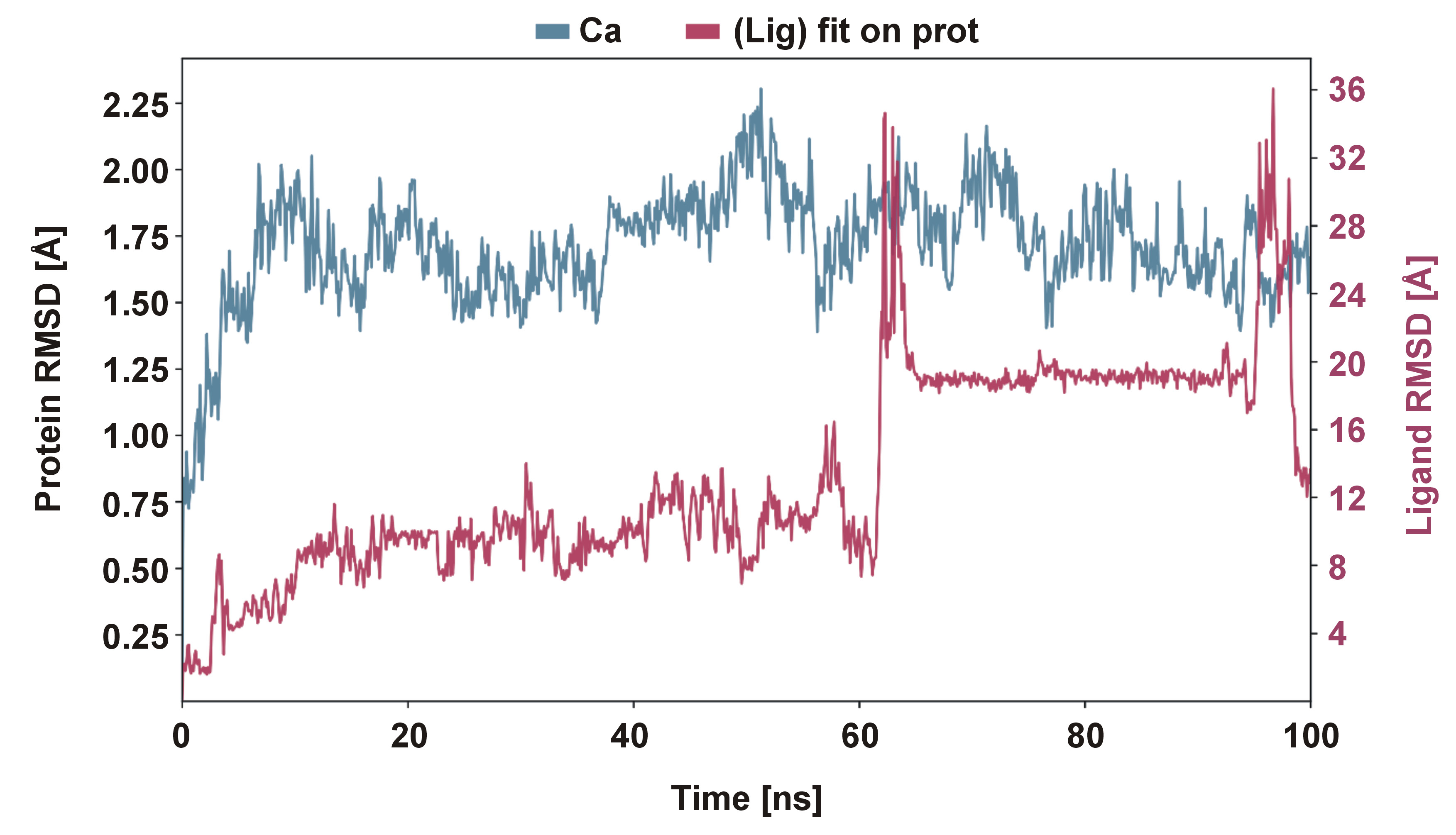
Results. During the course of the dynamic simulation, the trajectories were saved for the analysis every 100 ns. The stability of the complex was confirmed by a root mean square deviation (RMSD) plot. In the context of molecular docking, the protein (Lys-gingipain) and the ligand (4-CQA) were found to have a potential binding site with the use of hydrogen bonds. The compound had a docking score of −6.6 kcal/mol. According to the results of the dynamic study, as depicted in the RMSD plot, the compound demonstrated stability within the range of 1.0–3.0 Å.
Conclusions. The inhibition of Lys-gingipain by 4-CQA is a promising avenue for further investigation, whether in vitro or in vivo.
Keywords: Moringa oleifera, chronic periodontitis, molecular docking, gingipains, Porphyromonas gingivalis
Introduction
Periodontitis is a common oral condition prevalent in individuals with poor oral hygiene and affecting both the hard and soft supporting structures of the teeth. Additional factors that could influence the course of the disease include various systemic conditions, stress, malocclusion, orthodontic therapy, ill-fitting dentures, and genetics.1 Population-based studies have concluded that untreated periodontitis results in compromised quality of life due to functional, aesthetic and social disabilities.2 Periodontitis is caused by dysbiosis of oral microbial flora colonizing the supra- and subgingival regions of the teeth.3 Regarding the polymicrobial etiology, the complex red organisms are commonly associated with periodontitis, including Porphyromonas gingivalis, Treponema denticola and Tannerella forsythia.3, 4 Of P. gingivalis, a Gram-negative anaerobic rod exhibits a more significant effect on the oral microbiome and disrupts host–microbe hemostasis.5 As P. gingivalis initiates the dysbiosis of oral flora, it is called the keystone pathogen. It establishes and propagates periodontal disease through a virulence factor called gingipain. Gingipains are a group of cysteine proteases that are found in the outer membrane or the vesicles of the bacteria.6 They play a crucial part in maintaining the pathogenic functions of the bacterium in the host. Gingipains aid in colonization and adherence of the pathogen to epithelial cells, as well as cause the breakdown of erythrocytes, resulting in hemolysis. The generated heme provides an additional nutritional source for further multiplication and colonization of the bacteria. Subsequently, P. gingivalis modulates the host inflammatory response, leading to further progression and tissue destruction.7
Based on its amino acid composition, a gingipain can be classified as8:
• arginine-specific gingipain A (RgpA);
• arginine-specific gingipain B (RgpB);
• lysine-specific gingipain K (Kgp).
Lysine-specific gingipain K has a maximum of 3–5 hemagglutinin (HA)/adhesin domains in its genomic protein. In contrast, there are only 4 HA/adhesin domains in RgpA and no such domains in RgpB. As the number of domains increases, there is a concomitant increase in the effectiveness of host cell adhesion and heme acquisition.9 The distribution of domains in Kgp is a critical virulent factor of P. gingivalis.8 The analysis of the pathogens involved in the development of this pathology is a recurrent theme in the literature, which underscores its importance as a public health problem. Precisely for this purpose, the ability to identify targeted treatments against selected bacterial species is of current and future interest.10
Scientific evidence has suggested that since P. gingivalis is the keystone pathogen of periodontitis, treatment strategies directed towards it may prevent disharmony of the oral microbiome and inhibit disease progression. There are several treatment modalities for periodontitis, including non-surgical therapy involving scaling and root planning with or without antimicrobial therapy to decrease the microbial load, host modulation therapy, and surgical therapy for repairing or regenerating the lost periodontium.11 Advanced treatment strategies that could inhibit gingipain functions may have an indirect biological effect on P. gingivalis. They could suppress the availability of micronutrients for the growth of the bacterium and make it vulnerable to the defensive actions of immune cells. Apart from chemotherapeutic agents, natural remedies such as tulsi, aloe vera, neem, propolis, tea tree oil,12 and tropical fruits like mangosteen13 have been studied for their anti-microbial effect in the non-surgical treatment of periodontitis. The extensive research on herbal remedies is attributable to their anti-inflammatory, antioxidant and antimicrobial properties, along with a reduced incidence of side effects.14
Moringa oleifera, also known as the miracle tree, is one of the most commonly cultivated trees in tropical and subtropical regions. It is a rich source of phytonutrients, vitamins, minerals, and essential amino acids. Moringa oleifera also provides a rare combination of zeatin, quercetin, sitosterol, caffeoylquinic acid, and kaempferol.15 Various in vitro studies have concluded that many parts of M. oleifera, like leaves, pods, barks, nuts, flowers, and tubers, possess significant health benefits.16 4-Caffeoylquinic acid (4-CQA) is a phenylpropanoid of M. oleifera, the bioactivities of which include antioxidant, antibacterial, antidiabetic, anticancer, and antihistaminic effects.17 The antibacterial activity of phenolic compounds has been attributed to the loss of cellular integrity, leading to increased membrane permeability, subsequent disturbance of the cellular membrane, and ultimately, cell death.18
Molecular docking is a computer-assisted tool that is used to identify the complete binding site between the target protein and the drug in a three-dimensional assembly.19 This identification is a preliminary step in drug design that is performed before conducting in vitro and in vivo research. Hence, the present study aimed to assess the potential inhibition of P. gingivalis Lys-gingipain by 4-CQA of M. oleifera by means of in silico docking and dynamic simulations.
Material and methods
Molecular docking and dynamic simulations of the Lys-gingipain protein and 4-CQA ligands20 were performed using the Desmond software (Schrödinger, New York, USA).
The 3D protein structure of Lys-gingipain was downloaded from the Protein Data Bank (PDB) database (https://www.rcsb.org/structure/3M1H). The protein structure was processed using the Maestro platform (https://www.schrodinger.com/platform/products/maestro; Schrödinger). The Protein Preparation Wizard (Maestro; Schrödinger) was used to preprocess the receptor–ligand complex. In general, water molecules present in the protein can be easily displaced by the ligand or be a hindrance to the binding pocket. Therefore, the preprocessing steps in the protein preparation removed all heteroatoms and loosely bound water molecules by the addition of hydrogen ions. The selected protein structure was then examined for gaps and built further to fill the loops. After optimization, it was minimized using the optimized potentials for liquid simulations (OPLS 2005) force field. Conformers for each compound were obtained using force field estimates by the OPLS 2005 between atoms within and between the molecules.
The ligand was prepared after downloading the chemical structure of 4-CQA from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/#query=9798666). Then, the ligand was subjected to energy minimization using the OPLS 2005 force field to achieve correct bond length, order and angle with minimal energy. This grid-based ligand docking method was used to evaluate the interaction between the protein and the ligand. A grid box was utilized to describe the protein’s binding site with the following dimensions: center X – 9.599; center Y – 3.6929; center Z – 29.3808; size of X – 50.2525510788; size of Y – 33.8531600094; and size of Z – 34.8833085537.
Molecular dynamic simulations
The System Builder tool was used in the preparation of each system. The tool is designed to simulate the protein and the ligand. The default solvent water model TIP3P (3 points of transferable intermolecular interaction potential) with an orthorhombic box was selected to perform the dynamic simulation. Since there is a water-mediated interaction between the protein and the ligand in this TIP3 model, water must be included during the docking calculation. The OPLS 2005 force field was utilized in the simulation to generate the essential topology records.
The addition of counterions served to neutralize the models. To simulate the physiological condition, 0.15 M of sodium chloride (NaCl) was added. The isothermal–isobaric (NPT) ensemble was used, maintaining a temperature of 300 K and 1-Atm pressure throughout the simulation. The models were loosened before the simulation. Every 100 ns, the trajectories were saved for the analysis. The stability of the simulation was confirmed by contrasting the root mean square deviation (RMSD) of the protein and ligand over time.
Results
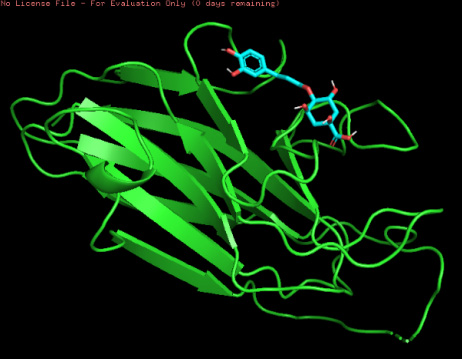
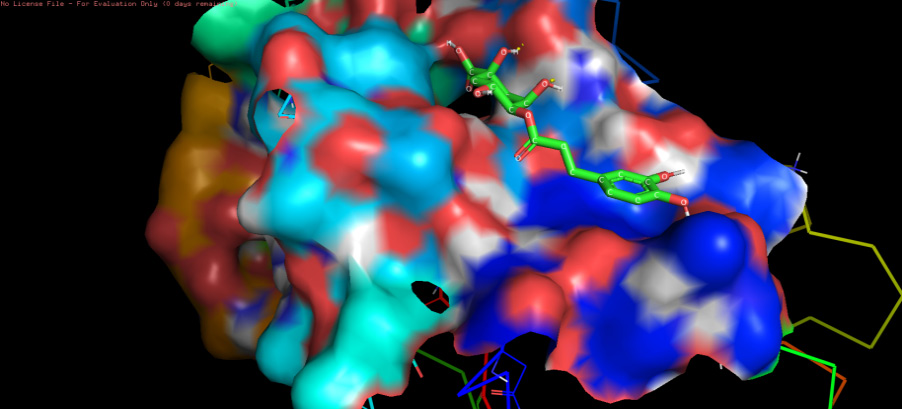
The three-dimensional docking model of the ligand–protein complex and the protein–ligand complex of Lys-gingipain and 4-CQA are depicted in Figure 1 and Figure 2, respectively.
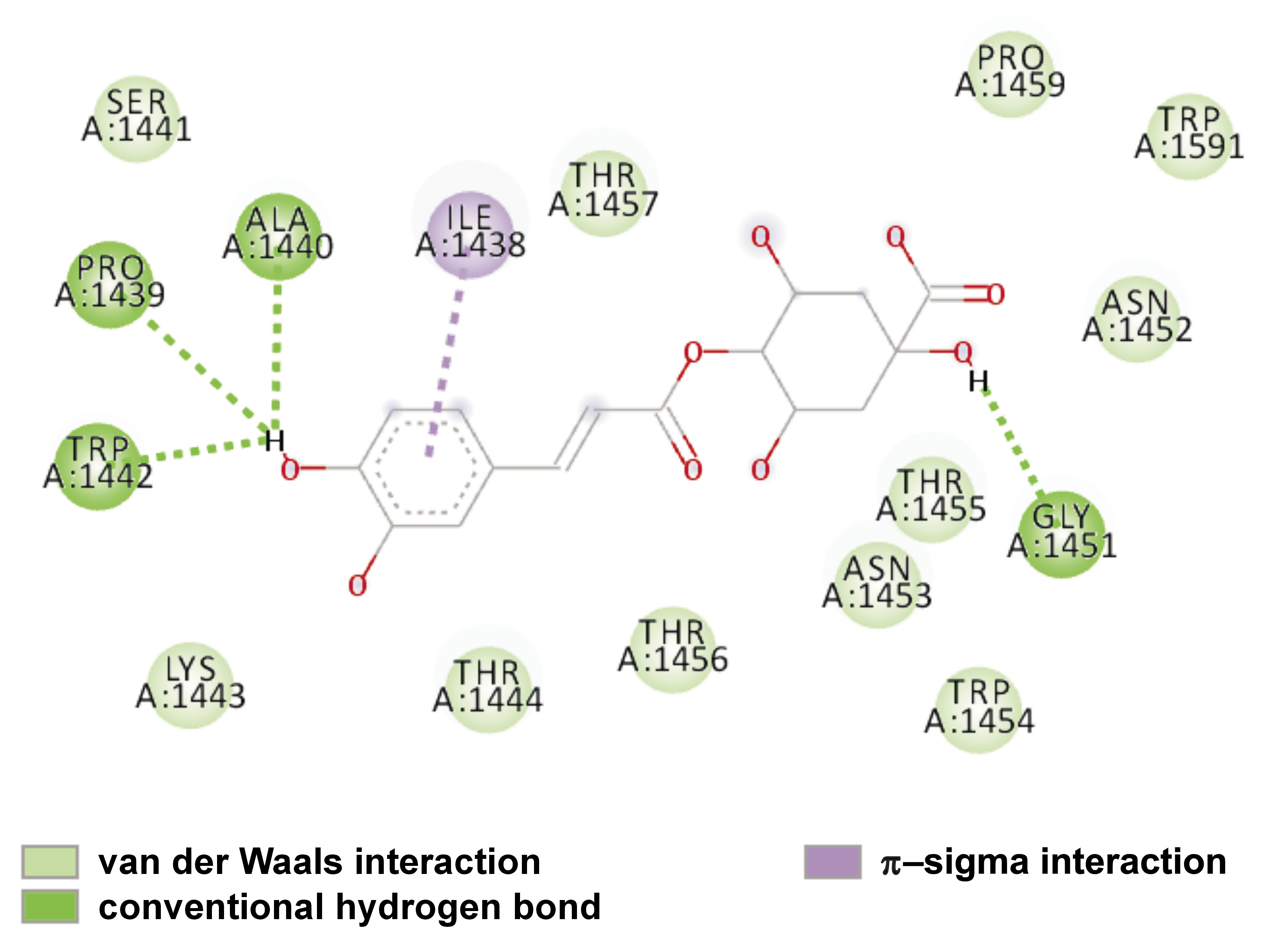
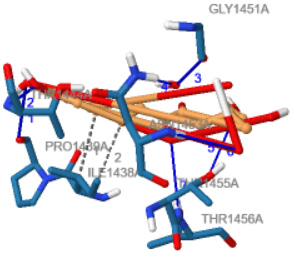
The interaction between the binding site residues of Lys-gingipain protein and the 4-CQA ligand are depicted two-dimensionally in Figure 3 and three-dimensionally in Figure 4.
The docking scores or binding affinities provide an estimation of the strength of the interaction between the ligand and the receptor. Lower scores or more negative binding energies generally indicate stronger binding. However, it is essential to validate these outcomes with experimental data or alternative methods to assess their reliability. Docking studies are a method of predicting the binding mode of a ligand within a receptor’s active site. An examination of the specific interactions, such as hydrogen bonding, hydrophobic contacts and electrostatic interactions, can help understand the key molecular interactions responsible for ligand binding. In the present study, the compound demonstrated a docking score of −6.6 kcal/mol, therefore substantiating strong binding between the ligand and the protein.
These results indicate that the 4-CQA ligand demonstrates the most favorable affinity for binding with the Lys-gingipain protein by forming strong hydrogen bond interactions with amino acids such as ALA 1440, PRO 1439, TRP 1442, and GLY 1451. The compound also forms a π–π interaction with ILE 1438.
Figure 5 presents the RMSD of the ligand molecule docked on the protein. According to the complex protein–ligand RMSD plot, the complex reached stability at 60 ns. Following this, the RMSD values for the protein remained within the 1.0–2.0 Å range throughout the simulation, while the ligand RMSD fluctuated within the 1.0–3.0 Å range. The RMSD figure demonstrated that the proteins within the complex reached stability at 60 ns. Following that, fluctuations in the RMSD values for the protein remained within the 1.0–2.0 Å range throughout the simulation. In contrast, the ligand RMSD values varied more widely, within the 8.0–36.0 Å range up to 100 ns, indicating higher conformational flexibility of the ligand within the binding pocket.
Table 1 presents the hydrophobic interactions between the amino acid residues of the protein and the ligand. Table 2 shows the hydrogen bonds formed between the amino acid residues of the protein and the ligand. These parameters include the interaction distances and the donor angles that characterize the bonding between the protein and the ligand.
Discussion
Periodontitis is caused by microbial pathogens present in biofilm complexes.21 These pathogens have a strong affinity to enter the bloodstream and rupture atherosclerotic plaque, causing fatality like stroke.22 Among the pathogens, P. gingivalis in the red complex group significantly contributes to chronic periodontitis.23 Though there are many periopathogens, P. gingivalis is the one that is commonly isolated from subgingival plaque samples.24 It has various virulence factors like fimbriae, capsules, vesicles, and proteases in the outer membrane.25, 26, 27 The virulence factors help colonize and multiply the bacteria by evading the host defense mechanism. Notably, heme is required for the growth of P. gingivalis, which, in turn, is acquired by the bacteria itself through the process of hemolysis of the host erythrocytes. The process of hemolysis is facilitated by the gingipain protease of P. gingivalis.28 Gingipains are a group of cysteine proteinases that are commonly found in the outer membrane of bacteria. Apart from degrading the proteins for their nutrition, they also compromise the host immune response and cause destruction of the periodontal soft and hard tissues.26 A periodontal treatment that targets the HA domain of gingipain has the potential to inhibit the colonization of P. gingivalis on the root surface.29 Cysteine peptidases, such as Kgp, RgpA and RgpB, which account for 85% of the pathogen’s extracellular proteolytic activity, are good candidates for inactivation. FA-70C1 is a strong P. gingivalis gingipain inhibitor derived from Streptomyces FA-70 culture supernatant.30 Previous research has reported the high-resolution (1.20) complex structure of Kgp with KYT-36, a peptide-derived, effective, bioavailable, and highly selective inhibitor.31
Phytocomponents are a rich source of protease inhibitors. In this regard, the extract of rice grain of Oryza sativa exhibited a significant gingipain inhibitory effect on Kgp and Rgps.30 These rice proteins were denoted as 16 unassigned peptidase inhibitor homologues in the database.30 Canavanine present in sword bean (Canavalia gladiata), when administered orally, decreased the alveolar bone loss in P. gingivalis-induced periodontitis in rats.32 Polyphenols present in cranberry significantly reduced biofilm formation by P. gingivalis and Fusobacterium nucleatum. Catechin, a polyphenol present in green tea, reduced the inflammatory reaction through its inhibitory effect on Rgp gingipain.33 Moringa oleifera contains phytocompounds, including flavonoids and phenolic acids. Phenolic acids are secondary plant metabolites that contain one or more hydroxyl groups connected to aromatic rings and act as scavengers.34 Among the phenolic compounds present in M. oleifera, CQA is also isolated from extracts of M. oleifera.35 Caffeoylquinic acid has shown antioxidant, antibacterial and anti-inflammatory properties.36
Caffeoylquinic acid is otherwise called chlorogenic acid (CA).37 A study was conducted to assess the effect of coffee on periodontitis, given the presence of caffeine and CA in the beverage.38 The study concluded that CA demonstrated significant anti-inflammatory activity because it inhibited nuclear factor kappa B (NF-κB) and resulted in substantial radical oxygen scavenging. The anti-inflammatory effect of CQA is enhanced in the presence of other phytocomponents, such as flavonoids.38
In another study, the antibacterial effect of CA was tested against P. gingivalis. The results demonstrated that CQA exhibited substantial anti-proteinase activity and had a prolonged inhibitory effect on P. gingivalis.39
The effect of 4-CQA of M. oleifera on Lys-gingipain has not been elucidated. The current study focused on the impact of CQA in M. oleifera on proteinase Lys-gingipain of P. gingivalis by in silico docking and dynamic simulation study. Based on the present findings, 4-CQA, a phenolic extract of M. oleifera, has been demonstrated to be a potent inhibitor of Lys-gingipain from P. gingivalis. The protein and the ligand exhibited a strong binding affinity for each other, as well as for amino acid residues like ALA 1440, PRO 1439, TRP 1442, and GLY 1451 by forming a hydrogen bond with the hydroxyl groups of the aromatic rings. The compound exhibited a docking score of −6.6 kcal/mol. According to the results of the RMSD plot, the compound demonstrated stability at 60 ns within the 1.0–3.0 Å range.
In subsequent in vivo studies, M. oleifera extracts can be applied topically to the periodontal pockets in the form of fibers, thermo-reversible gels, chips, or nanoparticle mouthwashes40 to test their efficacy against periodontal pathogens.
Limitations
The present study was able to assess the interaction between the protein and the ligand in an in silico molecular docking model. However, further research is necessary to confirm the bioactivity between P. gingivalis and 4-CQA in an in vitro design. Additionally, given that the blind docking technique was employed in this stage, future studies may utilize active site docking to evaluate the specific protein binding efficiency.
Conclusions
In recent years, ethnomedicine has emerged as a novel approach to prevent multi-drug resistance while treating various diseases, including periodontitis. This study concludes that 4-CQA, a phenolic extract acquired from the leaves of M. oleifera, could be used as a potent, novel and natural inhibitor of Lys-gingipain to prevent the progression of periodontitis.
Ethics approval and consent to participate
Not applicable.
Data availability
The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
Consent for publication
Not applicable.
Use of AI and AI-assisted technologies
Not applicable.