Abstract
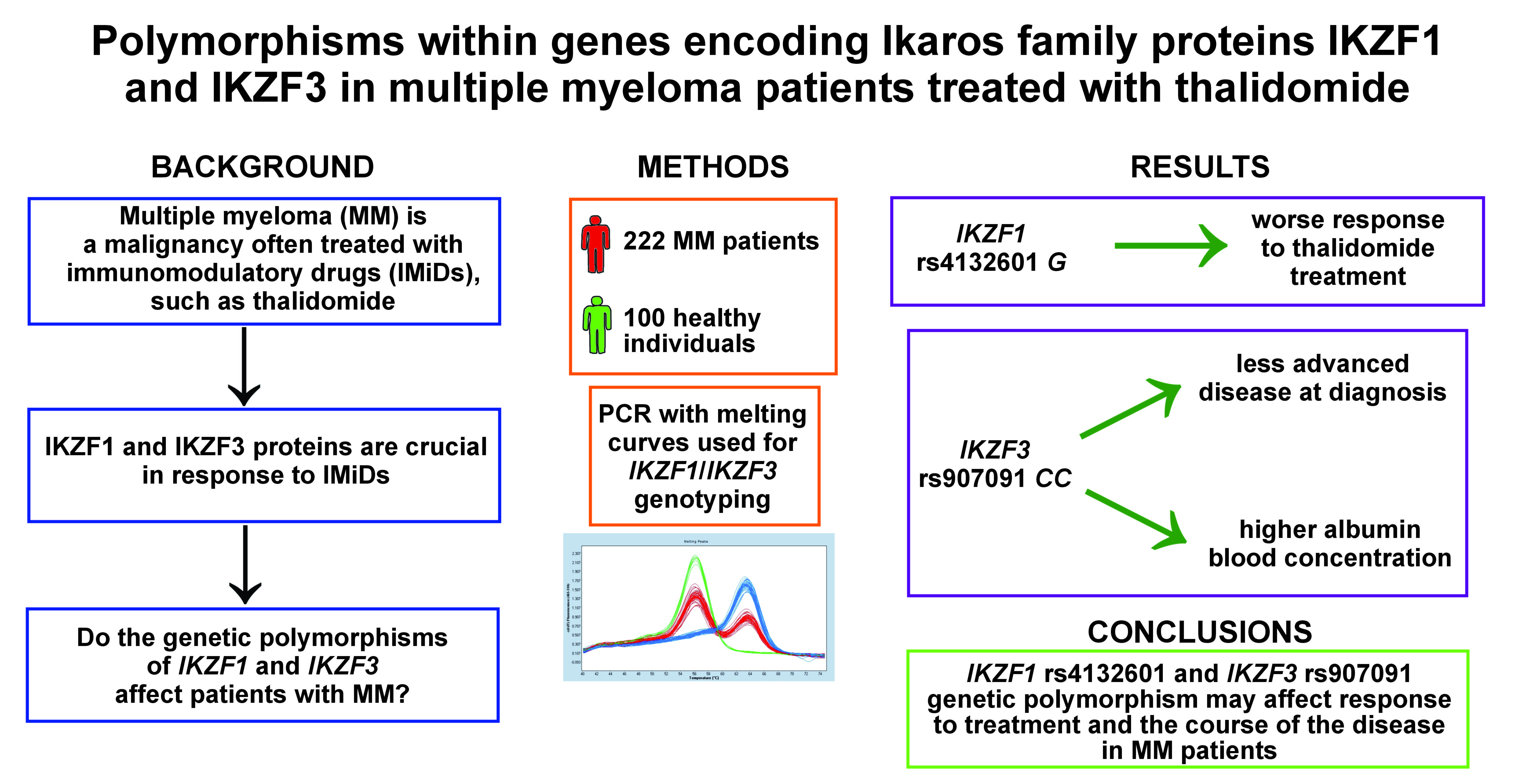
Background. Multiple myeloma (MM) is a hematological malignancy characterized by the presence of abnormal plasma cells. It is associated with anemia, bone lesions and renal dysfunction. Immunomodulatory drugs (IMiDs) are commonly used in MM treatment. Recent studies indicate that their therapeutic effect is caused by binding to cereblon (CRBN), which in turn causes the degradation of 2 important immune cell regulatory factors, IKZF1 and IKZF3. These are necessary for the anti-myeloma effect of IMiDs. Their expression level has been shown to affect MM survival and response to treatment. Potentially important single-nucleotide polymorphisms (SNPs) in the genes coding for IKZF1 and IKZF3 have been identified, but they have not been analyzed in MM patients before.
Objectives. The study was designed to establish the relationship between 4 SNPs in the genes coding for IKZF1 (rs61731359, rs4132601 and rs10272724) and IKZF3 (rs907091), and MM survival, response to treatment and other parameters.
Material and methods. The study involved 222 MM patients, as well as 100 control individuals. The IKZF1 and IKZF3 genotypes were determined by the LightSNiP assay. Genotyping was performed in the real-time polymerase chain reaction (PCR) LightCycler 480 device.
Results. No difference was observed between the patients and the controls for any of the SNPs, but the IKZF1 and IKZF3 variants were associated with various clinical parameters. Allele IKZF1 rs4132601 G was more common in the patients with worse response to first-line therapy (p = 0.040), particularly in the patients treated with thalidomide (p = 0.017). The patients tended to have worse overall survival. IKZF3 rs907091 CC was detected more commonly in the patients in stage I than in those in stages II and III, according to the International Staging System (ISS) criteria (p = 0.015). This genotype was also associated with a higher albumin level (p = 0.033), and was less common in the patients with the albumin level below 3.5 g/dL (p = 0.030).
Conclusions. Our results suggest that IKZF1 rs4132601 and IKZF3 rs907091 may affect response to treatment and progression in patients with MM.
Keywords: single nucleotide polymorphism, multiple myeloma, Ikaros transcription factor
Introduction
Multiple myeloma (MM) is an incurable hematological malignancy, which is characterized by the monoclonal growth of abnormal cells derived from transformed plasma cells. Multiple myeloma is a progressive disease that evolves from monoclonal gammopathy of undetermined significance (MGUS) and smoldering multiple myeloma (SMM) into symptomatic MM.1 It is estimated that 35,730 new MM cases will be diagnosed in the United States in 2023 and 12,590 patients will die due to the disease.2 In the Polish population, MM accounts for 18% of all hematological malignancies, and over 2,000 new cases are reported annually.3 The disease is diagnosed mostly in elderly people, and the median age at diagnosis is 69 years.2 Multiple myeloma is associated with poor prognosis; however, new drugs have greatly improved 5-year overall survival and patients’ quality of life (QoL).4, 5
Multiple myeloma typically leads to and is associated with anemia, lytic bone lesions, hypercalcemia, and renal dysfunction.6 It is believed that both environmental and genetic factors are responsible for the development of MM.1 Drugs currently available for MM treatment include proteasome inhibitors, monoclonal antibodies, 3-phosphoinositoside kinase inhibitors, and immunomodulatory drugs (IMiDs). Immunomodulatory drugs are a group that comprises thalidomide and its derivatives – lenalidomide and pomalidomide. They exert their therapeutic effect by binding to cereblon (CRBN), a component of the E3 ubiquitin ligase complex.7, 8 By binding to CRBN, IMiDs change the substrate specificity of the E3 ubiquitin ligase complex, and cause it to ubiquitinate and target for proteasomal degradation two B cell transcription factors – Ikaros family zinc finger protein 1 (IKZF1, Ikaros) and Ikaros family zinc finger protein 3 (IKZF3, Aiolos). Ikaros family proteins are known to be the major regulators of immune cell development, particularly with regard to CD4+ T helper cells, as well as B cells during their early and late development.9, 10 IKZF1 acts as a tumor suppressor in normal cells, but in MM cells, it becomes a transcriptional activator of oncogenes and promotes cancer cell survival.11 The knockdown and overexpression of IKZF1 and IKZF3 reduces the sensitivity of MM cells to IMiDs.12, 13, 14 Both IKZF1 and IKZF3 are needed for IMiDs to work, and the inactivation of either of them leads to MM cell cycle arrest.15 The degradation of IKZF1 and IKZF3 may lead to MM cell apoptosis through the downregulation of interferon regulatory factor 4 (IRF4) and c-MYC. It has also been shown that IKZF1 and IKZF3 repress interferon-stimulated genes (ISGs), including CD38, the expression of which is increased upon IMiD treatment.
Earlier studies established that the IKZF1 expression levels affected response to therapy and survival in patients treated with IMiDs.16, 17, 18 However, as of yet, no studies have investigated whether genetic variability within the genes coding for IKZF1 and IKZF3 may affect survival or response to treatment in MM patients. Our previous studies showed that the single-nucleotide polymorphisms (SNPs) located in the genes associated with IMiD metabolism might correlate with response to treatment and survival in MM patients.19, 20, 21 In the present study, we aimed to analyze selected SNPs in the IKZF1 and IKZF3 genes in the context of MM. Three SNPs were in IKZF1 (rs61731359, a missense Asn>Asp substitution; rs4132601, a 3’ untranslated region (3’UTR) variant and a potential miRNA binding site; and rs10272724, located near IKZF1 and affecting survival in acute lymphoblastic leukemia) and one in IKZF3 (rs907091, a 3’UTR variant and a potential miRNA binding site).22, 23, 24, 25 The 4 SNPs were analyzed in relation to MM susceptibility, response to treatment, survival, and association with diagnostic and prognostic markers, such as albumin, beta-2-microglobulin (β2-M), C-reactive protein (CRP), or calcium (Ca).
Material and methods
Patients and controls
The study included 222 patients diagnosed with MM, 111 women and 111 men, aged 37–88 years (median age: 64 years). The only inclusion criterion for patients to enter the study was a diagnosis of MM. The exclusion criterion was a diagnosis of any other malignancy.
According to the International Staging System (ISS) criteria, 54 patients were in stage I, 67 in stage II and 81 in stage III.26 Detailed information on patients is presented in Table 1. In addition, 100 healthy blood donors (50 women and 50 men) served as a control group. All patients and control individuals were from the Lower Silesia region in western Poland, and were treated between 2015 and 2020. The study was performed in accordance with the Declaration of Helsinki, and received the approval of the Ethics Committee at Wroclaw Medical University, Poland (no. KB-297/2016).
Sample size
The sample size analysis was conducted based on one of the primary objectives of the study, assuming a lower occurrence of the IKZF1 rs4132601 G allele in patients with better response (complete or very good partial remission) to first-line therapy (effect size: 0.30). The minimum required sample size to detect this difference, assuming α = 5%, a power of 90% and a confidence level of 95%, was 184 patients in total. Additionally, a 10% risk of incomplete data was considered. The final minimum required sample size was 203 participants. The sample size analysis was performed using the G*Power software.27
Immunomodulating therapy
Regarding first-line therapy, 127 patients were treated with the thalidomide-containing regimens (thalidomide 100 mg/day), including 89 patients treated with cyclophosphamide, thalidomide and dexamethasone (CTD) up to 6 cycles, and the subsequent continuous thalidomide monotherapy. Of the rest of the patients, 76 were not treated with thalidomide, and 19 patients lacked data on thalidomide treatment. Response to first-line therapy was as follows: complete remission (CR) in 37 patients; very good partial remission (VGPR) in 29 patients; partial remission (PR) in 77 patients; minor remission (MR) in 11 patients; stable disease (SD) in 19 patients; and progressive disease (PD) in 12 patients. For the patients treated with the thalidomide-containing regimens, the response was as follows: CR in 24 patients; VGPR in 18 patients; PR in 49 patients; MR in 8 patients; SD in 15 patients; and PD in 8 patients. Response criteria are well described in a previous study.3
Genotyping
DNA was extracted from the samples of peripheral blood taken on ethylenediaminetetraacetic acid (EDTA), using Maxwell® 16 Blood DNA Purification Kit (Promega Corp., Madison, USA) or QIAamp DNA Blood Midi Kit (Qiagen, Hilden, Germany), following the manufacturers’ recommendations. The IKZF1 and IKZF3 genotypes were determined by the LightSNiP assay (TIB Molbiol, Berlin, Germany), which uses specific probes allowing to distinguish genotypes based on melting curves (melting curve genotyping). Genotyping was performed in the real-time polymerase chain reaction (PCR) LightCycler 480 device (Roche Diagnostics, Rotkreuz, Switzerland) on a 96-well plate. A negative control with no DNA was included. Conditions for PCR were as follows: 95°C for 10 min; 45 cycles of 95°C for 10 s, 60°C for 10 s and 72°C for 15 s. This was then followed by one cycle of 95°C for 30 s and 40°C for 2 min, and gradual melting from 75°C to 40°C. The genotypes were distinguished based on the melting temperatures of the samples and the shape of their melting curves.
Statistical analysis
The null hypothesis that there is no difference in the presence of the IKZF1/IKZF3 alleles between patients and controls was tested with Fisher’s exact test or the χ2 test, using the website http://vassarstats.net. The genotypes were tested for deviation from the Hardy–Weinberg equilibrium using the χ2 test. Survival was assessed with the Wilcoxon test and the Kaplan–Meier survival curves. Other associations were tested either with Fisher’s exact test or the Mann–Whitney U test. All of these tests, except for Fisher’s exact test and the χ2 test, were performed with the Real Statistics Resource Pack for Microsoft Excel (https://real-statistics.com/free-download/real-statistics-resource-pack). A p-value <0.05 was considered statistically significant.
Results
Distribution of the IKZF1 and IKZF3 alleles and genotypes among patients with multiple myeloma and control individuals
All patients and control individuals (control group) were genotyped for rs4132601, rs10272724 and rs907091, while 203 patients and all control individuals were genotyped for rs61731359. Details about the genotype and allele frequencies are presented in Table 2. Genotype frequencies for all 4 SNPs were in accordance with the Hardy–Weinberg equilibrium in both patients and controls. We detected no statistically significant differences between the patients and the controls in any of the 4 studied SNPs, in any of the 3 investigated models (dominant, recessive and additive) (Table 3). Regarding rs61731359A>G, only allele A was detected, as all the subjects carried genotype AA. Two IKZF1 SNPs, rs4132601 and rs10272724, were in very high linkage disequilibrium (r2 = 0.96, D’ = 0.98), prompting us to select only one of them for further analysis. Due to the reasons listed above, rs61731359 and rs10272724 were not included in further analysis.
Association of the IKZF1 and IKZF3 polymorphisms with response to immunomodulating therapy
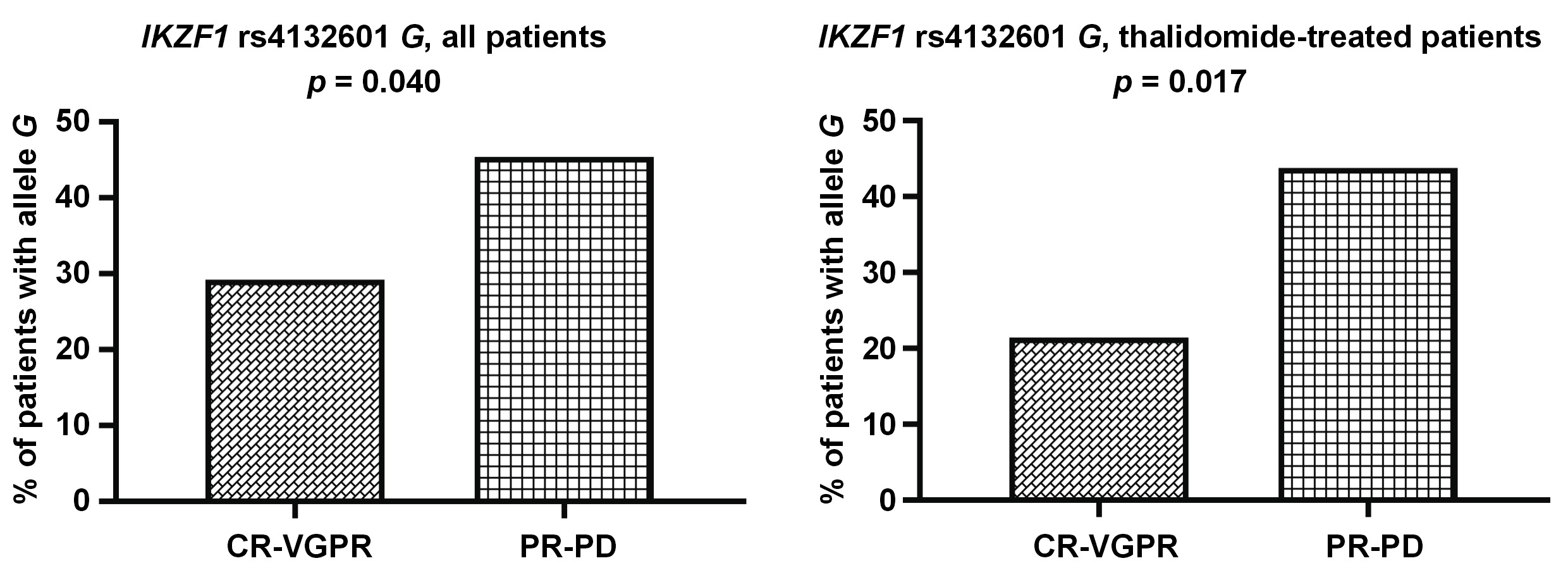
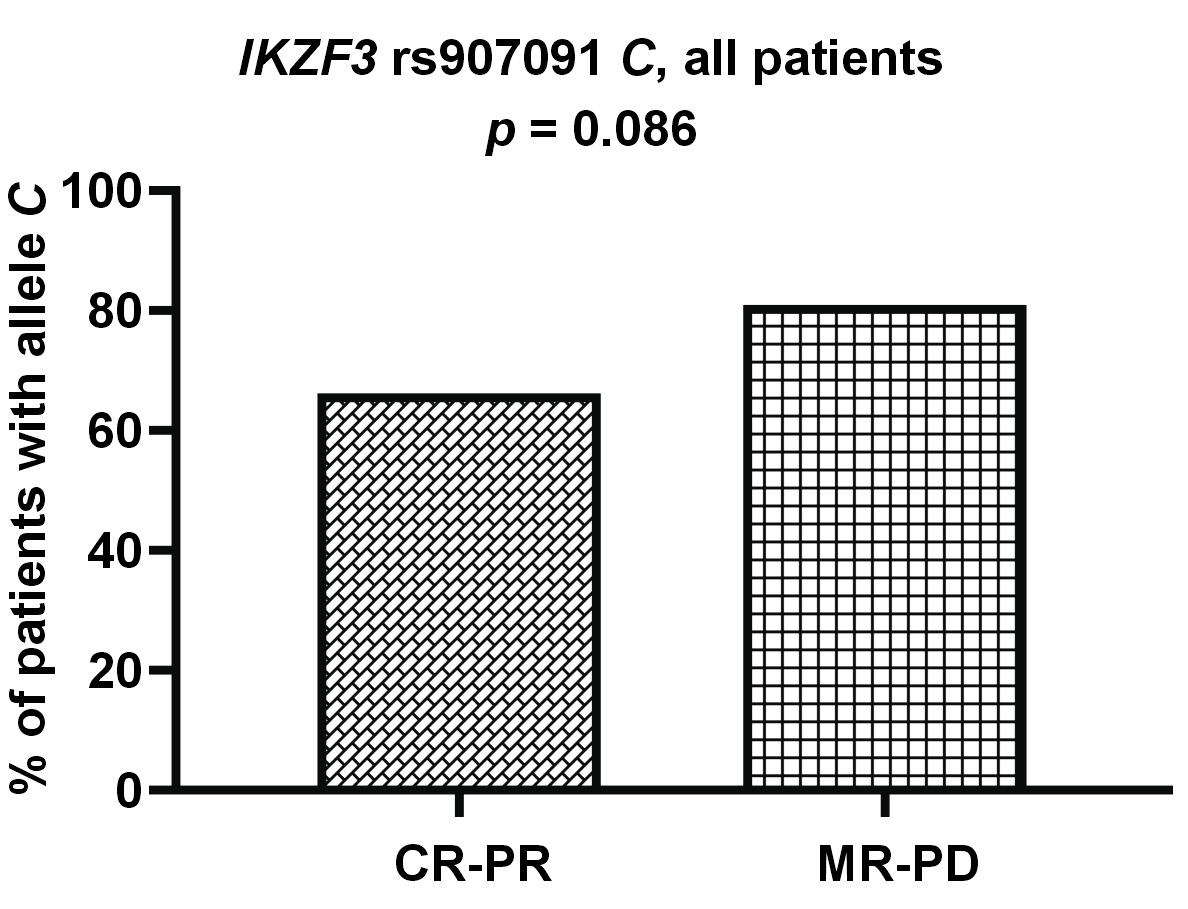
Allele IKZF1 rs4132601 G was less common among the patients with better response (CR-VGPR) to first-line therapy (19/66 vs. 54/119; p = 0.040). This association could also be observed in the subgroup of patients treated with thalidomide (9/42 vs. 35/80; p = 0.017) (Figure 1). Additionally, we observed a trend for worse response to treatment in the patients with allele IKZF3 rs907091 C, although this was not statistically significant (p = 0.086) (Figure 2).
Association of the IKZF1 and IKZF3 polymorphisms with survival and disease progression
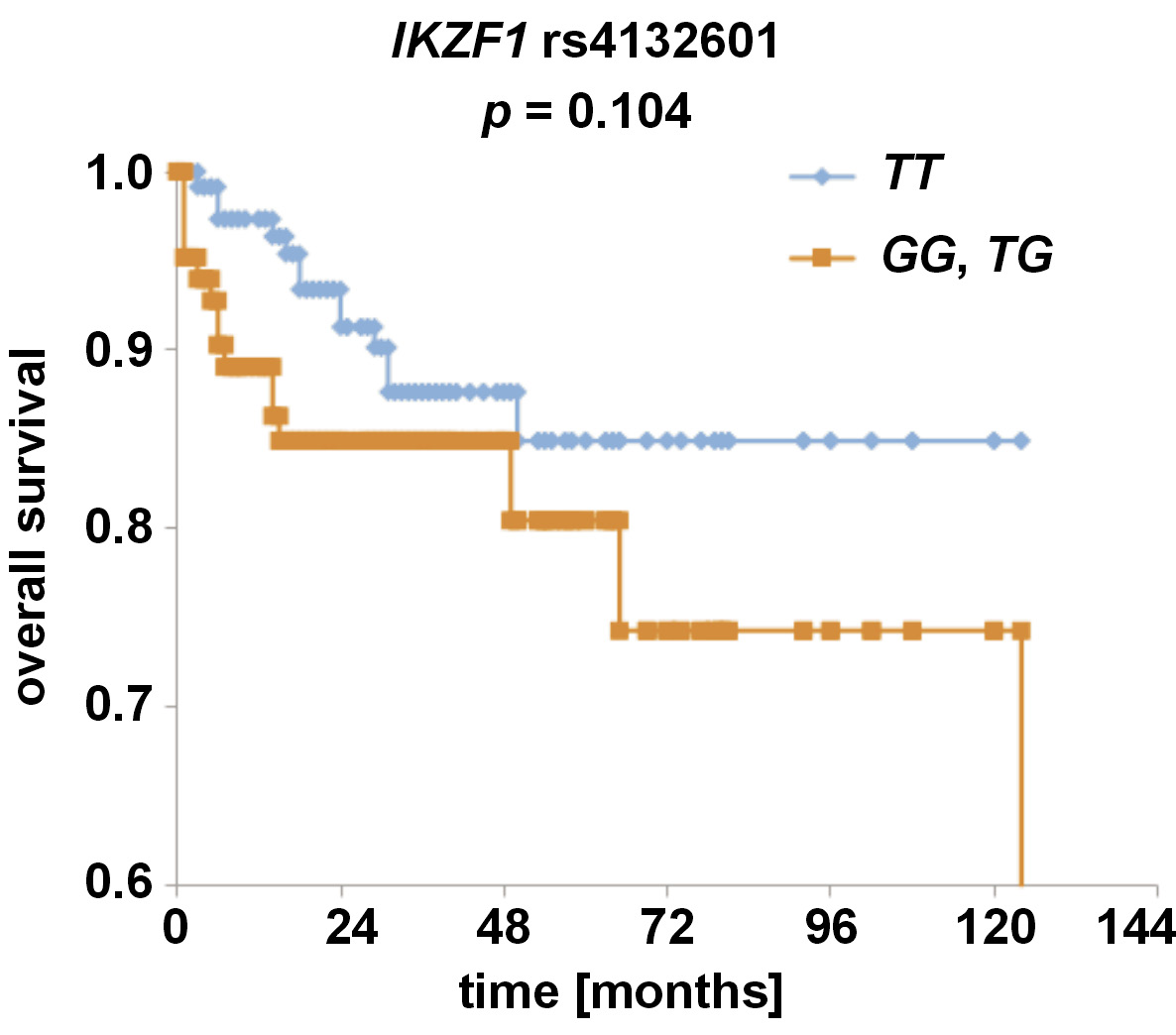
Overall and disease-free survival analyses were conducted for the IKZF1 and IKZF3 SNPs. While no statistically significant associations were observed for any of the analyzed SNPs, we noticed that the patients with allele IKZF1 rs4132601 G tended to have shorter overall survival than the patients without it (p = 0.104) (Figure 3).
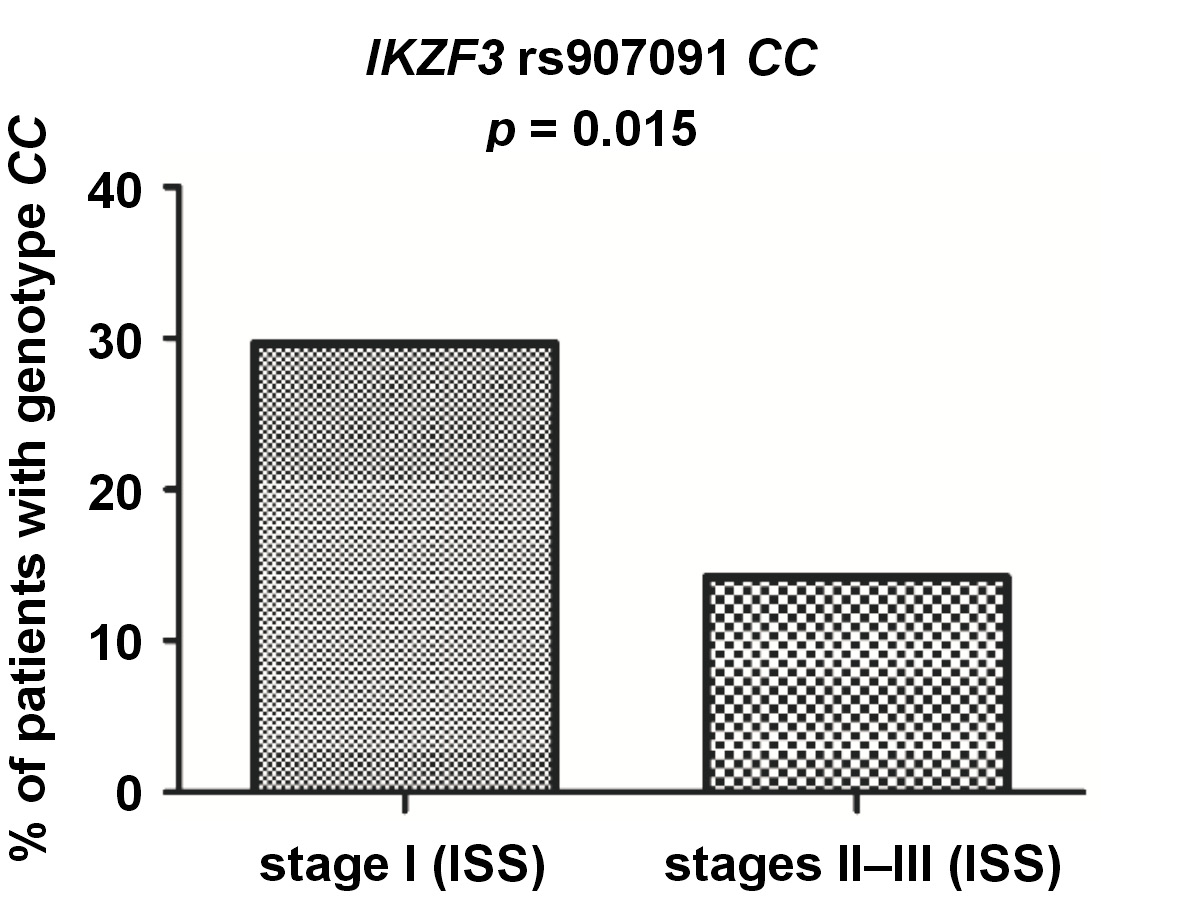
Regarding IKZF3, we found genotype rs907091 CC to be more common in the patients in stage I than in those in stages II and III, according to the ISS criteria (16/54 vs. 21/148; p = 0.015) (Figure 4).
Associations of the IKZF1 and IKZF3 polymorphisms with other prognostic factors
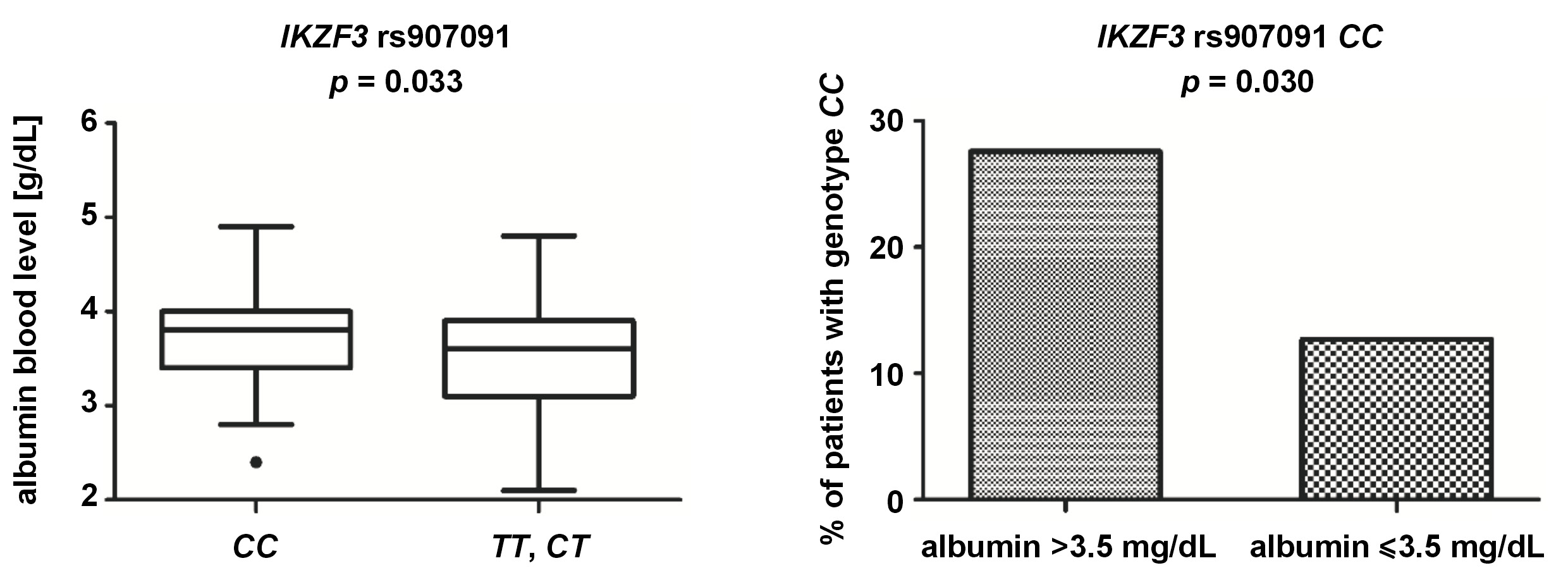
We observed that the median albumin blood concentration was slightly higher in the patients with genotype IKZF3 rs907091 CC than in the patients without this genotype (3.80 g/dL vs. 3.55 g/dL; p = 0.033). Furthermore, we noticed that CC homozygosity was less common in the patients with the albumin levels below 3.5 g/dL than in those with the albumin levels above 3.5 g/dL (9/71 vs. 24/86; p = 0.030) (Figure 5), with the albumin concentration below 3.5 g/dL being associated with worse prognosis.28 Additionally, the patients with genotype IKZF1 rs4132601 GG tended to have lower albumin concentration than the patients without this genotype (3.35 g/dL vs. 3.65 g/dL; p = 0.057).
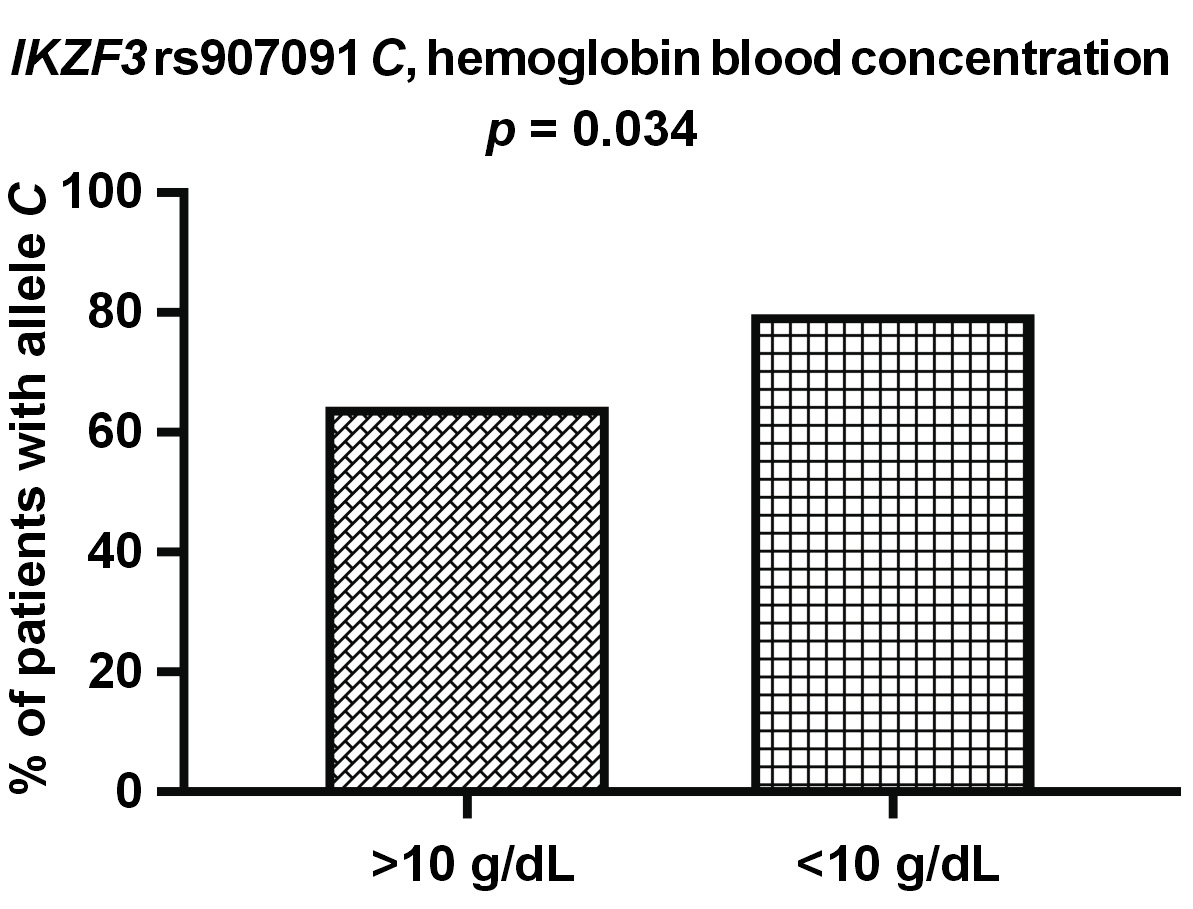
Regarding other clinical parameter – hemoglobin – allele IKZF3 rs907091 C was more commonly detected in the patients with abnormal hemoglobin blood levels (<10 g/dL) than in the patients with normal levels (55/69 vs. 81/126; p = 0.034) (Figure 6). We also observed that the patients with allele IKZF1 rs4132601 G tended to have higher Ca blood levels (9.53 mg/dL vs. 9.25 mg/dL; p = 0.082) and slightly higher age on diagnosis (65.0 years vs. 63.5 years; p = 0.053) than the patients without this allele, although this was not statistically significant. No associations regarding the levels of β2-M, creatinine and CRP, or the presence of light chains were detected.
Discussion
Immunomodulatory drugs are a major group of drugs used to treat MM. However, their mechanism of action has not been fully elucidated. It has been established that the molecular target of IMiDs is CRBN. While a ubiquitin ligase-independent pathway for IMiD action has been described, the anti-myeloma effect of IMiDs is most often ascribed to the degradation of IKZF1 and IKZF3 by the E3 ubiquitin ligase complex upon IMiD binding to CRBN.12, 13, 25 In the present study, we investigated the IKZF1 and IKZF3 genetic variants and analyzed their influence on survival, response to treatment and diagnostic markers in MM patients. Previous studies found differences in survival and response to treatment between patients with different IKZF1 expression levels,16, 17, 18 which may be caused by the interference of the genetic background. To the best of our knowledge, there has been no study analyzing the influence of SNPs in the genes coding for IKZF1 or IKZF3 on MM. Our study showed that allele rs4132601 G was associated with worse response to treatment, especially in patients treated with thalidomide, and might be more common in patients with shorter overall survival, although the latter association was not statistically significant. Furthermore, the G allele tended to be more common in patients with higher Ca blood levels, while the GG genotype tended to be more frequent in patients with lower albumin blood levels, which are associated with worse prognosis.25 However, it was not associated with susceptibility to MM.
IKZF1 rs4132601 was first identified as a risk variant in childhood acute lymphoblastic leukemia (ALL) in a study by Papaemmanuil et al,29 and has mostly been studied in the context of this disease. The researchers found the G allele to be associated with a higher risk for ALL and a lower IKZF1 mRNA expression level in a dose-dependent fashion.29 This association with the ALL risk was confirmed in various European, North African, Middle Eastern and Southeast Asian populations, although it was not detected in studies on ALL patients from Taiwan, Latvia and Iran.24, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39 The rs4132601 SNP was also analyzed in Polish diffuse large B cell lymphoma (DLBCL) patients.40 While the SNP was not associated with DLBCL susceptibility, allele G was found to correlate with longer progression-free and overall survival,40 contrary to our results in MM. This may be attributed to differences between the 2 diseases and the fact that DLBCL is not treated with IMiDs; therefore, the role of IKZF1 in this disease may be different from its role in MM. It is worth noting that our study showed that rs4132601 is in strong linkage disequilibrium with another IKZF1 SNP, rs10272724. It is thus possible that the effects ascribed to rs4132601 may, in fact, be due to a genetic variation in rs10272724.
Another SNP covered in our present study was rs907091, located in the gene coding for IKZF3, also known as Aiolos. Both IKZF1 and IKZF3 are needed for IMiDs to work, and the inactivation of either of them can lead to MM cell cycle arrest.15 We found rs907091 CC to be more common in MM patients with albumin levels above 3.5 g/dL (a marker of better prognosis) and to correlate with higher albumin levels in general. The rs907091 CC was also more commonly detected in patients in stage I of MM than in those in more advanced stages II and III, according to the ISS criteria. These findings may suggest that rs907091 CC is associated with better prognosis and slower progression of MM. On the other hand, we also observed that allele rs907091 C was more often detected in patients with abnormally low (<10 g/dL) hemoglobin levels. IKZF3 rs907091 was first described in a haplotype-based analysis that found it to be associated with gene expression changes.41 IKZF3 rs907091 is located in a predicted miR-326 recognition element (binding site) and miR-326 is expected to bind to rs907091 T (a variant absent in rs907091 CC homozygotes). Accordingly, patients with rs907091 T are reported to have lower IKZF3 expression.42 The rs907091 T was also been described to be associated with a higher risk for systemic lupus erythematosus (SLE) and multiple sclerosis (MS), with the CC genotype playing a protective role.42, 43, 44, 45 The protective effect of the CC genotype in these studies seems to correspond well with our results with regard to MM, although it should be noted that SLE and MS differ significantly from MM. No studies on IKZF3 rs907091 have been conducted in the context of MM or other hematologic malignancies.
Our study also included rs61731359, a functionally relevant IKZF1 SNP causing a missense Asn>Asp amino acid change.22 To the best of our knowledge, this variant has not been analyzed before in any group of patients. It did not have a polymorphic character in our group of MM patients and control individuals, as only one allele (A) was detected. This absence of allele G may have been due to the relatively small number of patients and controls investigated. Further studies on rs61731359 in MM and other diseases may be needed to elucidate its role.
Limitations
The relatively small number of patients and controls is a major limitation of our study. Another limitation that could have influenced our results is the non-homogeneous therapy used for MM treatment. Further studies on larger cohorts of patients are needed to confirm our results.
Conclusions
In conclusion, our present study shows that rs4132601 in the gene coding for IKZF1 and rs907091 in the gene coding for IKZF3 may be associated with response to treatment, survival, and the diagnostic and prognostic parameters in MM patients, such as the stage of the disease and albumin blood concentration.
Ethics approval and consent to participate
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Institutional Review Board of Wroclaw Medical University, Poland (approval No. KB-297/2016).
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Consent for publication
Not applicable.
Use of AI and AI-assisted technologies
Not applicable.