Abstract
Background. The activity of antioxidant enzymes in periodontitis is reduced, but results vary between studies and are subject to bias. In turn, the expression of genes encoding antioxidant factors has not been examined yet.
Objectives. This is the first study to evaluate the expression of genes encoding superoxide dismutase 1 (SOD1), glutathione peroxidase 1 (GPX1) and thioredoxin 1 (TXN1) in the saliva and gingival tissue of patients with periodontitis. The activity of the antioxidant enzyme protein products in the unstimulated and stimulated saliva and the gingival crevicular fluid (GCF) of patients with periodontitis was also investigated.
Material and methods. The prospective study involved 65 patients with periodontitis, who were divided into groups depending on the disease stage, and a control group of 31 age- and gender-matched healthy patients.
Results. We demonstrated that the expression of genes encoding GPX1 and TXN1 in saliva was significantly higher, and the expression of genes encoding SOD1, GPX1 and TXN1 in the gingival tissue was significantly lower in periodontitis patients as compared to the control group. We noted a lower activity of GPX1 in unstimulated saliva, of SOD1 in stimulated saliva and of both antioxidant enzymes in GCF in patients with periodontitis.
Conclusions. The GPX1 transcriptome and its activity in the salivary and GCF proteome appear to be dependent on the oxidative stress related to the destructive inflammatory changes in periodontitis.
Keywords: gene expression, saliva, periodontal disease, gingival crevicular fluid, salivary diagnostics
Introduction
During the course of periodontitis, due to the presence of a dysbiotic bacterial biofilm on the surface of dentin and cementum in the periodontal pocket, molecular pathways are activated in immune-inflammatory responses, leading to the destruction of the tooth-suspensory apparatus. Periodontitis is a social disease; in 2017, it was the 14th most common age-standardized diagnosis worldwide, with a prevalence of 9.8%, affecting approx. 796 million individuals worldwide.1 Periodontitis is an independent risk factor for selected systemic diseases with significant mortality, including diabetes, metabolic syndrome and cardiovascular diseases.2, 3, 4 Understanding the mechanisms of the protraction of inflammatory responses in periodontal tissues and the possibility of their effective interruption would be the basis for the primary prevention of the abovementioned diseases.
The periopathogen infection of the periodontal pocket stimulates a host immune-inflammatory response, in which neutrophils, T lymphocytes and B lymphocytes are activated. Neutrophils release reactive oxygen species (ROS) that directly damage periodontal tissues through lipid peroxidation, oxidative damage to proteins and DNA, the inhibition of cell growth, increased apoptosis, the destruction of the extracellular matrix (ECM) of gingival tissue and periodontal connective tissue, and elevated phosphatidylinositol activity.5, 6, 7 Activated T and B lymphocytes induce the production of receptor activator for nuclear factor-kappa B ligand (RANKL), which is an osteoclastogenetic mediator for the appendicular bone. The ROS signaling leads to the activation of the mitogen-activated protein kinase (MAPK), phosphoinositide 3-kinase (PI3K) and nuclear transcription factor nuclear factor-kappa B (NFκB) pathways.8 The NFκB signaling is the most important pro-transcription factor of numerous genes encoding pro-inflammatory cytokines, chemokines, adhesion molecules, and key enzymes for the synthesis of pro-inflammatory factors.9, 10 ROS detoxification is carried out by either cytoprotective antioxidant enzymes that prevent ROS from reacting with biological compounds (superoxide dismutase (SOD), catalase (CAT), glutathione peroxidase (GPX), glutathione reductase (GR), and the thioredoxin (TXN) system) or non-enzymatic antioxidants that interrupt free radical reactions (among others, glutathione (GSH), ascorbate, tocopherol, uric acid, and coenzyme Q). The transcriptional regulators of antioxidant enzyme genes are FoxO proteins (SOD2, CAT, GPX) and sirtuins (SOD2, CAT).8 Reactive oxygen species also activate cytoplasmic nuclear factor erythroid 2-related factor 2 (NRF2) through binding to Keap-1, resulting in its translocation to the nucleus and attachment to DNA, which initiates the transcription of genes that contain an antioxidant response element (ARE) sequence in the promoter. Those genes encode enzymes such as glutathione S-transferase, NADPH:quinone reductase, heme oxygenase, and γ-glutamylcysteine synthetase (γ-GCS).11 Increasing NRF2 activity may be a therapeutic target in periodontitis, leading to elevated local antioxidant activity and reduced pro-inflammatory signaling.
The analysis of the activity of antioxidant enzymes throughout the course of periodontitis has led to conflicting observations with regard to gingival tissue and gingival crevicular fluid (GCF).12, 13, 14, 15, 16, 17, 18 The evaluation of their activity in saliva previously indicated a significant decrease,17, 19, 20 although some authors described a significant increase.16, 21 Those differences can be explained by the intensity of the inflammatory process, its duration, the variety of research methods, the influence of periopathogens, or genetic conditions. It would be interesting to relate the expression of selected gene transcripts encoding antioxidant factors in gingival tissue and saliva to the concentrations of their protein products in GCF and saliva.
The present study aimed to evaluate the expression of SOD1, GPX1 and TXN1 at the mRNA level in gingival tissue and saliva, together with the activity of SOD1 and GPX1 in GCF and saliva in the most advanced stages and grades of periodontitis. Moreover, we investigated the covariability of mRNA expression of these genes and the activity of both antioxidant enzymes with clinical exponents of periodontitis.
Material and methods
Patients
The study was conducted in accordance with the STrengthening the Reporting of OBservational studies in Epidemiology (STROBE) statement. The protocol of the study was approved by the Bioethics Committee of Wroclaw Medical University (KB-559/2018). Informed written consent was obtained from all the subjects involved in the study.
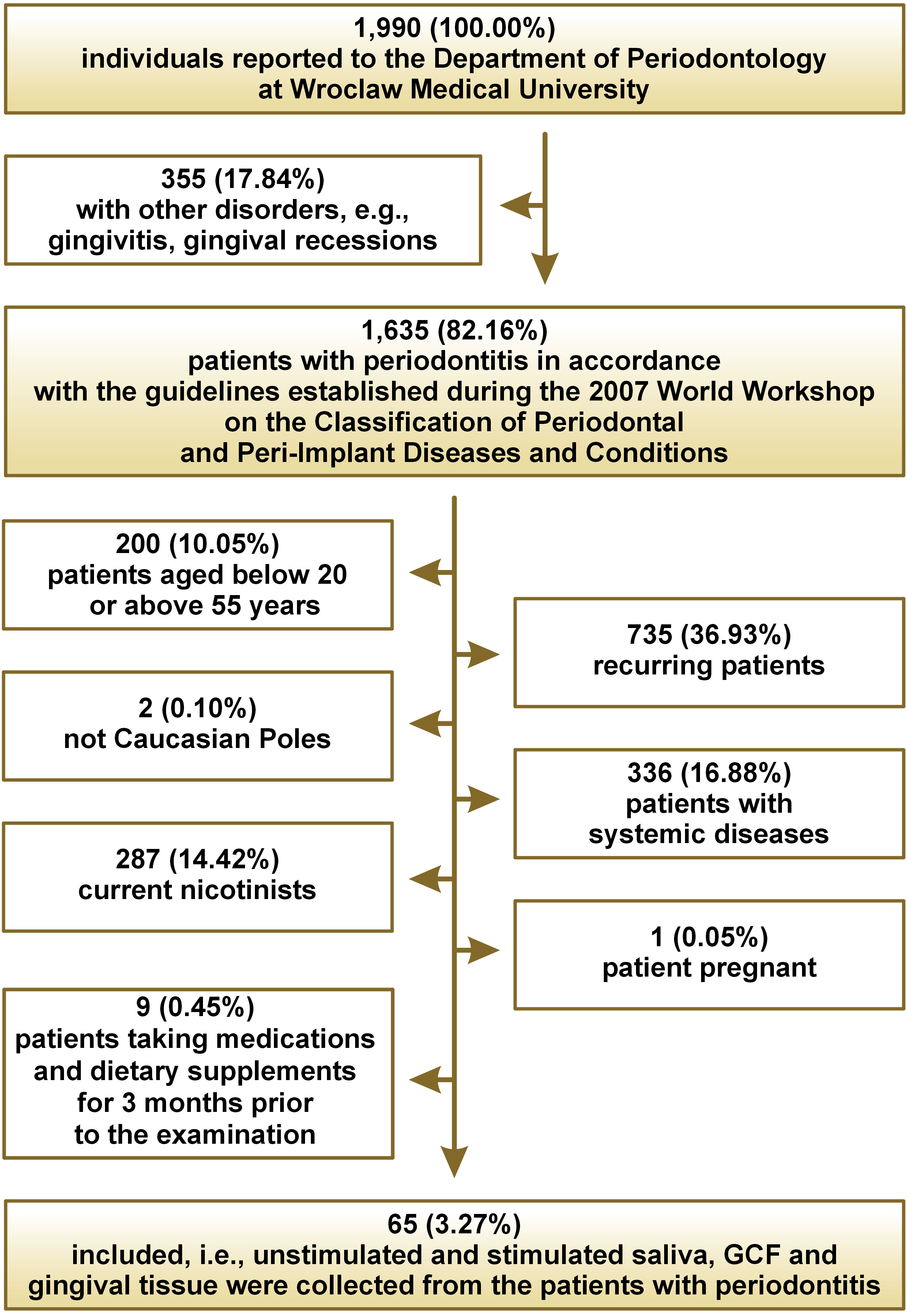
During the study period (January–December 2019), 1,990 patients reported to the Department of Periodontology at Wroclaw Medical University, Poland, of which 1,635 had periodontitis in accordance with the guidelines established during the 2017 World Workshop on the Classification of Periodontal and Peri-Implant Diseases and Conditions22 (Figure 1).
The exclusion criteria for both the study group and the control group were age below 20 and above 55 years, pregnancy, systemic comorbidities associated with oxidative stress (cancers, diabetes, hypertension, rheumatoid arthritis, kidney diseases, lung diseases, thyroid diseases), smoking in the 10 years preceding the study, the use of any medications or supplements in the 3 months preceding the study, the number of teeth below 15, the occurrence of clinical lesions in the oral cavity mucosa, or periodontal treatment less than a year before the study.
Of the original patients, 65 (3.27%) were prospectively qualified for the study. People with periodontitis were divided into 2 subgroups based on the current clinical criteria, namely stage III or IV and stage B or C.23
The control group, selected by gender and age to match the study group, consisted of 31 generally healthy patients of the Academic Dental Polyclinic in Wroclaw, Poland, with clinically healthy periodontium (bleeding on probing (BOP) <10%, pocket depth (PD) ≤3 mm).
Unstimulated and stimulated saliva, GCF and gingival tissue were collected from all patients.
Saliva collection
To minimize the effect of the circadian rhythm on saliva secretion, the samples were collected between 8 a.m. and 10 a.m., with any additional stimuli eliminated. For 2 h before the examination, the patients refrained from oral hygiene procedures and the consumption of any food or beverages (excluding clean water). Saliva was collected in a sitting position, with the head slightly tilted downward, into a sterile Falcon® (DNA/RNA-free) tube placed in an ice container.24, 25 Saliva was collected using the spitting method after rinsing the patient’s mouth 3 times with distilled water at room temperature and discarding the saliva collected during the 1st minute. Unstimulated saliva was collected for 10 min to a maximum volume of 5 mL. After a 5-minute break, stimulated saliva was collected for 5 min following stimulation by applying 10 µL of 2% citric acid on the tip of the tongue every 30 s. The volume of saliva was measured using an automated pipette, with an accuracy to 0.1 mL. Immediately after saliva was collected, the samples were centrifuged at 5,000 × g for 20 min at 4°C,24, 25 and the supernatant fluid, to which an antioxidant (10 µL of 0.5 M butylated hydroxytoluene per 1 mL of saliva) was added, was preserved for testing. Such samples were frozen at –80°C.26 A 1-minute salivary flow, expressed in mL/min, was calculated by dividing the volume of saliva by the time necessary for its secretion.
GCF collection
The clinically deepest periodontal pockets were selected during the clinical examination. The region was isolated from saliva access with dental cotton rolls, and was then dried with compressed air. Gingival crevicular fluid was collected using PerioPaper Strips™, then an antioxidant (10 µL of 0.5 M butylated hydroxytoluene per 1 mL of GCF) was added and the samples were frozen at –80°C.26 Any strips contaminated with blood or saliva were discarded. To determine the volume of GCF before and after the collection of the material, the strips were placed in Eppendorf® tubes and weighed on an analytical balance.27, 28
Tissue collection
In the study group, gingival tissue was collected from the region of the deepest periodontal pocket at the time of periodontal treatment. The procedure was performed under topical anesthesia with articaine and adrenaline (Septanest, 1:100,000; Septodont, Saint-Maur-des-Fossés, France). According to the manufacturer’s instructions, the material was placed in RNAlater® tubes and frozen at –80°C. Gingival fragments without signs of inflammation were collected from the control patients at the time of a third molar extraction. In all patients, tooth extraction was performed only for orthodontic reasons.
Clinical trial
Immediately after unstimulated saliva, stimulated saliva, GCF, and gingival tissue were collected, each patient had the dental examination performed by the same dentist (TK) according to the criteria of the World Health Organization (WHO), namely under artificial lighting, with the use of a mirror, an explorer and a periodontal probe.29 The following clinical parameters were evaluated: the number of retained teeth; the mean mobility value of all teeth according to the indications of the Periotest® device; the modified plaque index (PI) according to O’Leary et al.17; bleeding on probing (BOP) by Ainamo and Bay18; the papilla bleeding index (PBI) by Saxer and Mühlemann19; and the pocket depth (PD) measured at 6 points of each tooth. Furthermore, the mean PD for all teeth, the mean interproximal PD measured at 4 points of each tooth, the number of sites with PD > 5 mm, and the clinical attachment level (CAL) measured at 6 points of each tooth were calculated.
RNA isolation
The evaluation of the mRNA expression of the genes obtained from gingival tissue and saliva was performed at the Department of Molecular Techniques at Wroclaw Medical University.
The RNA isolation from the saliva samples was performed using the ISOLATE Biofluids RNA Kit (Bioline, London, UK) according to the manufacturer’s instructions. Solid tissues were mechanically homogenized with the use of MagNA Lyser Green Beads (Roche Diagnostics, Mannheim, Germany) in Lysis Buffer and Homogenate Additive (Thermo Fisher Scientific, Inc., Waltham, USA). Subsequently, total RNA was extracted using the mirVana miRNA Isolation Kit (Thermo Fisher Scientific, Inc.), as recommended by the manufacturer. The RNA samples were then stored at –20°C.
RT-PCR
The cDNA synthesis was performed for each sample using the High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific, Inc.) with random hexamers and 10 µL or 14 µL of RNA isolated from solid tissues or saliva, respectively. Individual reactions were conducted in a total volume of 20 µL under the following thermal conditions: 25°C for 10 min; 37°C for 2 h; and 85°C for 5 min. The expression levels of GPX1, SOD1 and TXN were measured by means of the relative real-time polymerase chain reaction (RT-PCR) method, using the TaqMan™ Gene Expression Assays (GPX1: Hs00829989_gH; SOD1: Hs00533490_m1; TXN: Hs00828652_m; GAPDH: Hs99999905_m1) and the TaqMan™ Fast Universal Master Mix (Thermo Fisher Scientific, Inc.). All reactions were performed in triplicate in a total volume of 10 µL, using the 7900HT Fast Real-Time PCR System (Thermo Fisher Scientific, Inc.), under the following thermal cycling conditions: 95°C for 20 s; 40 cycles of 95°C for 1 s; and 60°C for 20 s. All results were normalized against the expression of GAPDH and calculated using the 2–ΔΔCt method.
Enzyme activity
The analysis of the activity of SOD1 and GPX1 in GCF and saliva was conducted in the Saliva Biochemistry Laboratory of the Department of Conservative Dentistry at the Medical University of Bialystok, Poland. All measurements were conducted using double tests and were standardized to milligram [mg] of total protein.
On the day of the measurements, the samples of saliva and GCF were slowly thawed at 4°C. To extract GCF, the PerioPaper Strips were placed in an Eppendorf test tube containing 0.02 M phosphate-buffered saline (PBS) solution with pH of 7.0 (1 strip/500 µL PBS). The samples were mixed for 30 s by using a vortex mixer, and then centrifuged at 3,000 × g for 5 min at 4°C. The supernatant fluid was preserved for testing.27, 28 An antioxidant (10 µL of 0.5 M butylated hydroxytoluene per 1 mL of GCF) was added to the samples containing GCF, and then they were mixed with a vortex mixer.30 Gingival crevicular fluid was used for all measurements on the same day. The saliva and GCF samples were mixed with a vortex mixer immediately before the measurements.
The activity of SOD1 (E.C. 1.15.1.1) was measured by means of the colorimetric method described by Misra and Fridovich.31 The principle of that method is based on the measurement of the cytoplasmic SOD activity subunit in the inhibition reaction of oxidation of epinephrine to adrenochrome at 320 nm. It was assumed that 1 unit of SOD activity inhibited 50% of epinephrine oxidation. The absorbance changes were measured at 320 nm. The SOD activity was measured in duplicate, and it was expressed in mU/mg of total protein.
The activity of GPX1 (E.C. 1.11.1.9) was evaluated colorimetrically, measuring the conversion of NADPH to NADP+ at 340 nm.32 One unit of GPX activity was defined as the amount of enzyme that can catalyze the oxidation of 1 mmol NADPH per 1 min. The GPX1 activity was measured in duplicate, and it was expressed in mU/mg of total protein.
Statistical analysis
The Mann–Whitney U test was used for assessing differences between the 2 groups, whereas the evaluation of 3 groups was conducted using the Kruskal–Wallis test, followed by post hoc Tukey’s test. Spearman’s test was used for the correlation analysis. Statistical significance was determined at p ≤ 0.05 for the Mann–Whitney and Kruskal–Wallis tests, while p ≤ 0.02 was considered statistically significant for the correlation analysis. The statistical package Statistica, v. 13.3 (StatSoft, Cracow, Poland), was used.
Results
The general and periodontal data of the patients are shown in Table 1.
mRNA data
The gingival SOD1 mRNA expression was significantly higher in controls as compared to all patients with periodontitis. It was also significantly lower in both stages of periodontitis as compared to controls, although the difference in expression was not statistically significant in the most advanced stage of the disease. The salivary SOD1 mRNA expression was not significantly different throughout the course of periodontitis and both its most advanced stages as compared to controls (Table 2).
The gingival GPX1 mRNA expression was also significantly higher in controls as compared to all patients with periodontitis. It was significantly lower in stage III periodontitis as compared to controls. In contrast, the difference between stage IV periodontitis and controls was not statistically significant. The salivary GPX1 mRNA expression was significantly higher throughout the course of periodontitis and both its most advanced stages as compared to controls, although it did not differ between the disease stages (Table 3).
The gingival TXN1 mRNA expression was significantly higher in controls as compared to all patients with periodontitis. It was also significantly lower in both stages of periodontitis as compared to controls. The salivary TXN1 mRNA expression was significantly higher during periodontitis progression and both its most advanced stages as compared to controls, although it did not differ between the disease stages (Table 4).
Enzyme activity
The activity of SOD1 in GCF was significantly lower in all patients with periodontitis as compared to controls. That observation was also true for stage IV periodontitis. Similarly, the SOD1 activity in unstimulated saliva was significantly lower in all study groups and in the patients with stage III periodontitis as compared to controls. There were even more clear differences in the SOD1 activity in stimulated saliva between the control group and the whole study group, as well as stage III and stage IV periodontitis (Table 5).
The activity of GPX1 in GCF was significantly lower in all patients with periodontitis as compared to controls, as well as in stage III and IV periodontitis. The GPX1 activity in unstimulated saliva was also significantly lower in all patients with periodontitis as compared to controls, and in stage III and IV periodontitis. The activity of GPX1 in stimulated saliva in all patients with periodontitis and its III and IV stages were, in turn, significantly higher as compared to controls (Table 6).
Clinical correlations
The analysis of the differences in the mRNA expression of the analyzed antioxidant genes, and in the activity of SOD1 and GPX1 in GCF and saliva between periodontitis with rapid and moderate progression rates demonstrated significantly higher SOD1 activity in stimulated saliva in grade C (Table 7).
The analysis of the covariation of the mRNA expression of the 3 analyzed genes between gingival tissue and saliva revealed no significant relationships. The same applied to the correlations between the mRNA expression of SOD1 and GPX1 and the levels of their SOD1 and GPX1 products in GCF and both types of saliva (data not shown). A significant positive correlation between the SOD1 mRNA expression in gingival tissue and the SOD1 activity in stimulated saliva in periodontitis patients with the fastest disease progression was the only exception (R = 0.63; p = 0.002).
The evaluation of the covariation of the SOD1 mRNA expression as well as the SOD1 activity and clinical parameters in all patients with periodontitis showed no significant correlations. In the case of gingival GPX1 mRNA expression, significant positive correlations were found with inflammation intensity (BOP: R = 0.31; p = 0.019) and the mean PD (R = 0.37; p = 0.005). The GPX1 activity in GCF and both types of saliva did not correlate with clinical parameters. The salivary TXN1 mRNA expression displayed a significant correlation with the number of periodontal pockets >5 mm (R = 0.32; p = 0.013). In the most advanced stage of periodontitis, the following significant correlations between antioxidant and clinical parameters were observed: the salivary SOD1 mRNA expression vs. BOP (R = 0.56; p = 0.006) and PBI (R = 0.51; p = 0.013); the salivary GPX1 mRNA expression vs. PBI (R = 0.51; p = 0.019); the GPX1 activity in stimulated saliva vs. the percentage of teeth with CAL ≥ 5 mm on the interproximal surfaces (R = –0.61; p = 0.016); the gingival TXN1 mRNA expression vs. the mean PD (R = –0.54; p = 0.007) and the number of periodontal pockets >5 mm (R = –0.57; p = 0.005); and the salivary TXN1 mRNA expression vs. the number of sites with CAL > 5 mm (R = 0.54; p = 0.007). Only 4 statistically significant correlations were observed in patients with the fastest progression of periodontitis, namely the salivary SOD1 mRNA expression vs. BOP (R = 0.53; p = 0.004) and PBI (R = 0.47; p = 0.013); the gingival GPX1 mRNA expression vs. PBI (R = 0.46; p = 0.013); and the salivary TXN1 mRNA expression vs. PI (R = 0.45; p = 0.013).
Discussion
The present study indicates that at the mRNA level of SOD1 expression in the gingiva of patients with periodontitis, there is downregulation as compared to the clinically healthy periodontium. During the course of periodontitis (especially in its most advanced stage), the enzymatic activity of that protein was also reduced in GCF and both unstimulated and stimulated saliva. In the most rapidly progressive stage of periodontitis, a significant positive covariation was observed between SOD1 mRNA expression in the gingiva and SOD1 activity in unstimulated saliva, as well as a significantly higher SOD1 activity in this type of saliva. Under the conditions of ROS and reactive nitrogen species (RNS) formation during periodontitis-related oxidative and nitrosative stress, there is an intranuclear downregulation of SOD1 expression through the NRF2 signaling pathway.33 The post-translational mechanisms of SOD1 regulation, namely phosphorylation, lysine modification and S-acetylation, also play an important role in its enzymatic activity.34 The post-translational modification of histones through the epigenetic mechanisms, stimulated by periopathogens could also alter the expression of many genes, including antioxidant genes. However, one study showed no significant differences in the methylation of CpG sites of the SOD1 gene in gingival epithelial cells between patients with periodontitis and controls.35 The protraction of the inflammatory process in periodontal tissues leads to the enzymatic depletion of ROS scavengers, as shown by the evaluation of their activity, particularly in GCF.36 In addition to the observations presented herein, a significant reduction of SOD1 activity in GCF during periodontitis has also been described by other authors.17, 18, 37 Periopathogens play a special role in the processes taking place in the periodontal pockets. Sampath et al. demonstrated that Porphyromonas gingivalis-infected cells showed a significant elevation in the GSK-3β, and a reduction in NRF and SOD1 mRNA expression as compared to uninfected cells.38 Although the expression of the salivary mRNA transcript of SOD1 did not differ between periodontally ill and healthy individuals in the present study, the salivary activity of that enzyme was markedly reduced during the course of periodontitis. The former part of the above observation has not been previously published, while the latter one is consistent with the results of other authors.17, 19, 20 However, opposite results have also been reported.16, 21 These discrepancies are likely due to several determinants, namely the phase of periodontal tissue inflammation, its duration, the sources of salivary antioxidants other than those related to the periodontal pockets, the number and composition of bacteria in saliva, the short half-life of SOD1, the oral hygiene procedures, and methodological considerations.39 The SOD1 activity in stimulated saliva was the only antioxidant parameter analyzed that stratified the degrees of periodontitis. However, that was probably due to the outflow of blood from the periodontal pockets into saliva at the most advanced stage and grade of periodontitis, as indicated by significant positive correlations of salivary SOD1 mRNA expression with the extent and intensity of the inflammatory response.
While the SOD1 transcription appears to be independent of the direct influence of periodontitis, the GPX1 RNA transcription may depend on the intensity of the destructive inflammatory process within periodontal tissues. In the present study, this is indicated by the significant positive correlations of the GPX1 transcript levels with the intensity of gingival inflammation and periodontal pocket depths, as well as of the expression of this transcript in stage C disease with the inflammation intensity in the gingiva in all periodontitis patients. The expression of GPX1 depends not only on ROS, but also on selenium availability, an inflammatory response to antigen stimulation and insulin resistance.40 Through kinase phosphorylation pathways (c-Jun N-terminal kinase (JNK), extracellular signal-regulated kinase (ERK), MAPK, and others), ROS activate FoxO proteins, while elevated nuclear levels of FoxA1 are a factor that regulates the transcription of the GPX1 gene encoding the cytoplasmic GPX1.41 In the present study, the gingival GPX1 mRNA expression was significantly reduced in all patients with periodontitis and those suffering from stage III periodontitis as compared to controls. This is not consistent with the observation made by Duarte et al., who found a significantly higher gingival GPX1 mRNA expression in 15 generally healthy patients with periodontitis as compared to 12 controls.42 Those differences may be due to the greater severity of periodontitis in relation to the present study (our patients suffered from stage IV periodontitis and the difference in the expression of this transcript was blurred as compared to controls) and different study group sizes. The depletion of the efficiency of the enzymatic antioxidant mechanisms during the long-term course of periodontitis is also indicated by our observation of a significantly lower GPX1 activity in GCF as compared to controls. The literature references for that observation range from those finding no difference to those showing a significantly higher activity throughout the course of periodontitis.43, 44 However, our results are supported by a reference showing the levels of oxidized glutathione in GCF, indicating a significant reduction in the mean GSH levels in GCF from the periodontal pockets as compared to clinically healthy sites.45, 46 This may be compounded by the mechanism of scavenging the bactericidal hypochlorous acid through adjusting the biosynthesis of intracellular glutathione in Porphyromonas gingivalis-infected gingival epithelial cells toward a phenotype that promotes periopathogen survival in the periodontal pockets.47 Our analysis showed that the salivary GPX1 mRNA expression and the GPX1 activity in stimulated saliva in all patients with periodontitis and those suffering from the 2 most advanced stages of the disease were significantly elevated, whereas they were reduced in unstimulated saliva as compared to controls. This confirms the assumption that the source of salivary RNA is not only the outflow of fluid and blood from the periodontal pocket (the positive correlation between the GPX1 mRNA expression and PBI in the most advanced stage of the disease), but also the 3 major salivary glands, minor salivary glands, serum transudate in the salivary glands, and the exfoliating cells of the parakeratotic epithelium of the oral cavity.48 The expression of salivary RNA with a short half-life is further influenced by a diverse oral microbiome, with significant differences in the content and types of endo- and exoribonucleases. Certainly, this may limit the sensitivity and specificity of such studies, although a strong pro-inflammatory stimulus in the form of extensive periodontitis can significantly alter the salivary transcriptome. The stimulation of saliva secretion provides additional extra-periodontal antioxidant factors, hence there are differences in the activity of enzymatic antioxidants between the evaluated types of saliva.49 However, in other studies, the evaluation of the GPX1 activity in saliva was again differential, with unstimulated saliva showing a significant reduction or elevation,21, 43, 50, 51 while stimulated saliva demonstrated a reduction.19
A pioneering element of the current study was the evaluation of the TXN1 mRNA expression in the gingiva and saliva of patients with periodontitis. Unfortunately, due to insufficient material for the study, the levels of TXN1 in GCF and saliva were not determined. There was a significant reduction in the gingival TXN1 mRNA expression both in the whole group of periodontitis patients and separately for the 2 most advanced stages of the disease. Moreover, in stage IV periodontitis, there was a significant negative correlation of that expression with the mean PD and the number of periodontal pockets deeper than 5 mm, although this does not confirm the dependence of this expression on the periodontitis progression. Oxidative stress leads to the dissociation of the Keap-1–NRF2 complex and the entry of NRF2 into the cell nucleus, where it binds to the adenylate-uridylate-rich elements (AREs) of DNA in the promoter regions of TXN1 on chromosome 9q31.3 and TXNRD1 on chromosome 12q23-q24.1.52 Also, in the case of this enzymatic antioxidant system, this process is successively reduced with the long-term course of periodontopathy. In addition to ROS and RNS, other factors that enhance the expression of these genes include ultraviolet (UV) radiation, retinoic acid, selenium availability (TRXD1 is a selenoreductase), and reoxygenation after hypoxia. In the GCF extracted from healthy periodontal sites, a protein containing TXN domains was confirmed among 199 identified proteins following albumin depletion.53 Through the spectrometric method of the matrix-assisted laser desorption/ionization (MALDI) with a tandem time-of-flight (TOF) analyzer (MALDI-TOF), TXN1 was found in the GCF extracted from the periodontal pockets.54 After the use of liquid chromatography with tandem mass spectrometry (LC-MS), a higher expression was found in GCF from the periodontal pockets as compared to gingival clefts.55 Herein, a significantly higher salivary TXN1 mRNA expression was found in periodontitis patients and its most advanced stages as compared to controls. This is likely due not only to the secretion of that transcript into saliva from the periodontal pockets (positive covariations with the number of periodontal pockets deeper than 5 mm in all periodontitis patients and the number of periodontal pockets with CAL above 5 mm in stage IV periodontitis), but also the product of glandular tissue of major and minor salivary glands,56 blood serum filtrate, and epithelial cell exfoliation. Thioredoxin was found with the use of a quadrupole mass spectrometer (QMS) and a TOF analyzer, while thioredoxin peroxidase was observed in whole saliva by using MALDI-TOF.57 A study of the proteome of unstimulated whole saliva (UWS) with the use of LC-MS significantly more frequently found TXN1 in the whole saliva of periodontitis patients as compared to periodontally healthy individuals, and also observed a significantly higher intensity of its expression during periodontitis and its close functional interactivity with catalase.58 Interestingly, Lee et al. observed significant negative correlations between increases in the bleeding extent/PD and the TXN1 levels in stimulated saliva in individuals with untreated periodontitis (who had a dental check-up frequency of less than once a year).59 Such correlations were not observed in patients with regular dental check-ups.59 This shows the possibility of depletion of the TXN antioxidant system with the disease progression, and it may also involve other antioxidant enzymes. In our patients, who had the rapidly progressive form of periodontitis, a positive covariation was observed between the salivary TXN1 mRNA expression and an indicator of the presence of a bacterial biofilm visible on the tooth surfaces. However, other authors do not confirm this relationship for the TXN1 expression in saliva.58, 59 Perhaps this is not a coincidental relationship, but caused by a specific potentiation of the activity of the TXN system against the oxidative stress induced by the periopathogen Fusobacterium nuclaetum, which is the most important element in supragingival biofilm formation.60
Conclusions
In conclusion, during periodontitis as defined by the current trends, there is a significant reduction in the mRNA tissue expression of the SOD1, GPX1 and, to the greatest extent, TXN1 genes, as well as a significant elevation in the salivary expression of transcripts of the GPX1 and TXN1 genes. Surprisingly, patients with stage IV periodontitis were more similar to healthy controls than patients with stage III. This may be due to the induction of the antioxidant defense mechanisms, which increase with disease progression. It is well known that the enhancement of the antioxidant barrier is the primary adaptive mechanism to prevent oxidative damage in the oral cavity. In the case of 2 transcripts of gingival oxidative genes, no correlation with the activity of their protein products in GCF and saliva was observed. The GPX mRNA expression and the GPX activity in GCF and saliva appear to be most dependent on the oxidative stress related to the destructive inflammatory changes in periodontitis. It does not appear that any of the examined antioxidant elements could act as a predictor of the degree of periodontitis.
Further studies of antioxidant parameters in patients with periodontitis may lead to the elucidation of prognostic and diagnostic factors, and create concepts of new therapeutic strategies.
Ethics approval and consent to participate
The protocol of the study was approved by the Bioethics Committee of Wroclaw Medical University (KB-559/2018).Informed written consent was obtained from all the subjects involved in the study.
Data availability
The article contains complete data used to support the findings of the present study.
Consent for publication
Not applicable.